IMAGING & IMAGE PROCESSING
The non-invasive monitoring of tissue engineered constructs in an environment that closely mimics in vivo conditions is one of the main challenges in regenerative medicine. In fact, the ability to visualize cells seeded on a scaffold on encapsuled in spheroids, as well as self-assembled in organoids, can help elucidate cell/biomaterial interactions without sacrificing samples.
Brain Imaging
The study of brain micro-architecture is of interest for the investigation of disease related alterations in neural structure, for the study of the so-called connectome and for quantitatively measuring the degree of similarity with the human in vivo counterpart. Despite the enormous efforts directed to the study of the brain’s microstructure and its correlation with higher order functions, the lack of protocol standardization and optimization makes it difficult to reproduce inter-lab data, which often leads to misleading conclusions and conflicting results. Moreover, the presence of lipids in brain tissue limits light penetration depth due to scattering and constitutes a barrier for high-resolution 3D imaging of intact brain tissue. In addition, algorithms for neuron segmentation from 3D imaging datasets and for the extraction of morphometric variables from single neurons in their native arrangement within the brain are lacking, hindering the construction of connectome datasets to be shared within the scientific community.
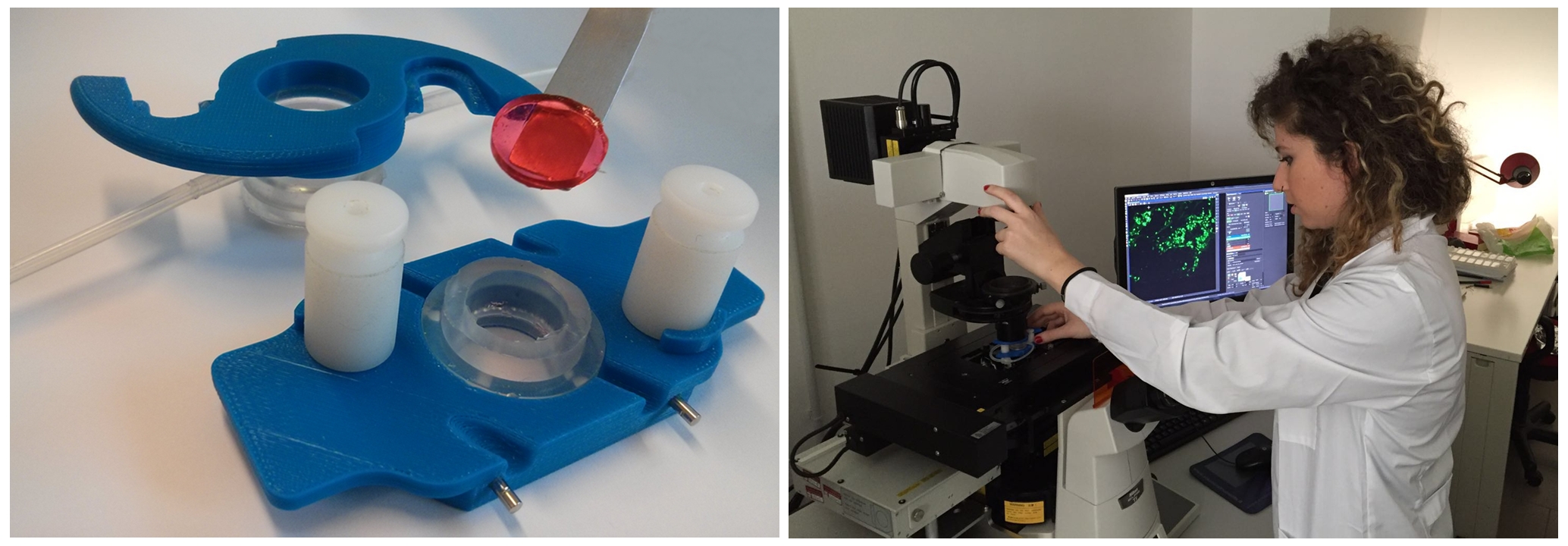
Our group extensively works on a systematic methodology for treating brain tissues and extracting quantitative information from 3D images.The methodology is based on three steps:
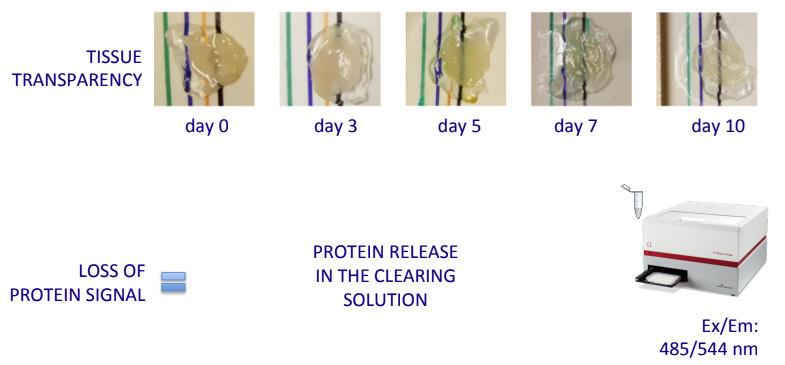
i) the optimization of tissue delipidation procedures
,
ii) the acquisition of datasets from advanced imaging tools,
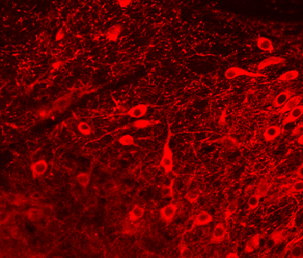
iii) the implementation of algorithms to automatically extract morphological features from the acquisitions.
The NanoBioscopy Lab
The NanoBioscopy Lab, jointly run by the Research Center E. Piaggio and the Department of Biology, is dedicated to advanced imaging of tissues for the study of physiological and disease models.
 Our publications
Our publications
 Contact info:
Contact info:

